By Yuanwei Xu and Hui Zhang, Center for Biomarker Discovery & Translation, Johns Hopkins University
Glycosylation is one of the most important protein modifications, playing an essential role in almost every aspect of biological processes. Despite being an important subdiscipline of proteomics, the investigations on glycoproteomics lagged behind not due to lack of interest, but a dearth of suitable methods for characterizing the tremendously complex glycoproteome. Luckily, we are able to characterize glycoproteome in an unprecedented depth with the help of advanced technologies. Glycoproteomics has come to the spotlight of completing the picture of human proteome.
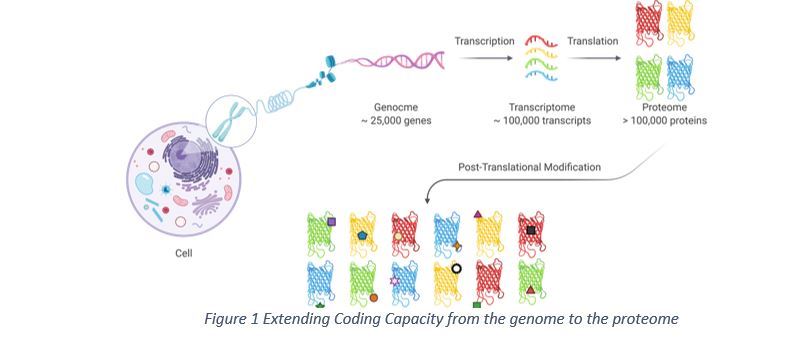
The Human Genome Project transforms biology and medicine through its integrated big science approach to decipher the roles of the human genome. The genome is almost identical across different human cells and throughout the life 1. However, the coded proteins from human genome in cells are much more dynamic and highly diversified to facilitate different biological functions2 3. In this context, the human proteome holds significantly more functional proteins than the coding capacity of 20,000 to 25,000 genes 1, which could be two to three orders of magnitude more complex (>1,000,000 protein species) 4 (Figure 1). The major mechanism responsible for the expansion of proteome is that proteins are subject to elaborative modifications 1 4.
Identifying proteins that are modified by specific chemical groups and determining their modification sites are the key focuses in characterization of human protein modifications. Ever since the 1980s 5, phosphorylation has been the most characterized protein modifications (based on the number of publications on the topic of different modifications in human from 1970 to 2020 at PubMed). Up to ~50,000 phosphorylation sites could be identified in each sample in a single phosphoproteomic experiment 6 7 8 9 10 11. Glycosylation, along with other modifications such as acetylation, ubiquitination and SUMOylation are more pervasively investigated because of technological advancements. Approximately, glycoproteins take up ~50% of the proteome 12, unmatched by any other protein modifications, since glycosylation is highly diversified to facilitate an assortment of functions 13. Despite the significance of protein glycosylation, the investigation of glycoproteome remains challenging due to the diversity of glycoprotein isoforms (glycoforms) when compared to other modifications. A rather comprehensive characterization of protein glycosylation site and the fine details of these site-specific glycans (including composition, sequence, branching, linkage, and anomericity) 14 would require for glycoprotein characterization.
Eukaryotic protein glycosylation is usually via two major types of linkages to proteins: N- and O-linked. N-linked glycoproteins are mainly attachment to asparagine residues by the covalent N-glycosidic bond. The general consensus peptide sequence for N-glycan is Asn-X-Ser/Thr (where X is any amino acid except proline) 15 16, while unusual glycosites with atypical motifis such as Asn-X-Val and Asn-X-Cys were found with low occupancy 17. Moreover, N-glycans in eukaryotic cells share a common core sequence, Manα1-3(Manα1-6)Manβ1-4GlcNAcβ1–4GlcNAcβ1–Asn 16. In contrast, O-glycosylation is linked to the hydroxyl groups of serine or threonine residues without an obvious motif preference 16. The initiating monosaccharides for O-linked glycosylation include galactosamine (GalNAc), mannose, galactose, fucose, glucose, and glucosamine (GlcNAc) 16, linear or branched oligosaccharide chains of various lengths could be further extended from the initiating monosaccharides. Both N- and O-linked glycosylation could be capped or modified with certain monosaccharides and chemical groups or substitutions 14. All of these aspects of glycosylation compounded protein glycosylation with a multitude of complexities. Other glycan-protein complexes form structures such as GPI-anchored glycoproteins and proteoglycans are also presented in eukaryotic systems.
Glycoproteomics focuses on the large-scale characterization of glycoproteins. Microarray-based approaches and mass spectrometry-based approaches have been used in glycoproteomics 14. Glycoprotein coding genes 18, purified glycoproteins 19, glycans 20 21 22 23, lectins 24 25 , or glycan-specific antibodies 26 have been used in microarray-based approach. Due to the plasticity of glycosylation, mass spectrometry (MS)-based glycoproteomics characterizes glycoproteins at different levels, including glycosylation sites (glycosites), glycans, and glycosite-specific glycans 27 28. Several enrichment methods have been published for these purposes, including hydrazide chemical tagging29 30, metabolic labeling 31 32, chemoenzymatic labeling 33, lectin chromatography 34 35 36, HILIC 37 38, ERLIC 39, “SimpleCell” technology with homogenized O-glycans 40 41 and EXoO42. The enriched glycosites, glycans, glycopeptides or glycoproteins would then be analyzed using different MS approaches, including CID-MS (often fragmenting glycans), ECD/ETD-MS (often cleaving peptide backbones), MALDI-MS (often for detailed glycan analysis using MSn), and HCD-MS (often generating both peptide backbone and glycan fragment ions). The MS results would then be searched against known databases to identify glycosylation sites, glycans, glycans at each glycosite, and the abundance of certain glycoforms at each glycosite. Up to June 2020, 14,644 unique N-glycosylation sites, 30,872 unique N-linked glycosite-containing peptides and 7,204 unique N-linked glycoproteins were identified in human (based on outputs at N-GlycositeAtlas 43: http://glycositeatlas.biomarkercenter.org/#). As for O-linked glycosylation, 4,672 unique O-glycosylation sites were identified across the human brain, kidney, T cells, and serum using EXoO 44. In total, 3,369 glycan structures (based on outputs at GlyCosmos Portal: https://glycosmos.org/glytoucans/list) are identified in human. Glycoproteomic databases are emerging, yet we are only beginning to see the tip of the iceberg.
Apart from developing a suitable MS-based analytical approach, advanced computing power is indispensable for the characterization of glycoproteins. Sophisticated algorithms have been developed into software to assist in the interpretation of mass spectra. SEQUEST 45 and the recently developed pFind 46 are dedicated for the high-throughput peptide and protein identification via tandem mass spectrometry. GlycoPep ID 47, GlycoPep DB 48, and GlycoMod 49 are some of the freely accessible web-based programs for glycopeptide analysis. Skyline 50 and MaxQuant 51 are frequently adopted for large-scale quantitative proteomic studies. GlycoWorkBench 52, SimGlycan 53, Cartoonist 54, and MultiGlycan 55 can be used for the interpretation of glycan spectra, UnicarbDB 56 57 holds one of the largest experimental MS/MS databases on released glycans, while Byonic 58, GPQuest 59, and pGlyco 60 61 are developed to analyze intact glycopeptide spectra.
The recent technology advancement has brought numerous innovative approaches in various aspects of analytical glycoproteomics, including sample preparation, enrichment, mass spectrometric analyses, data analysis tools, and databases. As a result, the complexities of glycosylation characterization are reduced to a large extend. Along with the growing attention placed upon the alterations of glycosylation in every biological perturbation, especially in pathological states, the glycosylation “enigma” has been unraveling at a faster pace. Being the center of glycomics, glycoproteomics finally comes to the spotlight of human proteome characterization.
References


.png)