|
HGI study 2committee membersCo-Chairs:
Stacy Malaker, PhD Yale University, New Haven, CT, USA
Nicholas M. Riley, PhD Stanford University, Palo Alto, CA, USA Committee members: Soka University, Tokyo, Japan
Frédérique Lisacek, PhD Swiss Institute of Bioinformatics, Geneva, Switzerland
Nichollas E. Scott, PhD University of Melbourne, VIC, Australia
Northeastern University, Boston, USA Ad hoc advisors:
Macquarie University, Sydney, Australia
Daniel Kolarich, PhD Griffith University, Gold Coast, Australia |
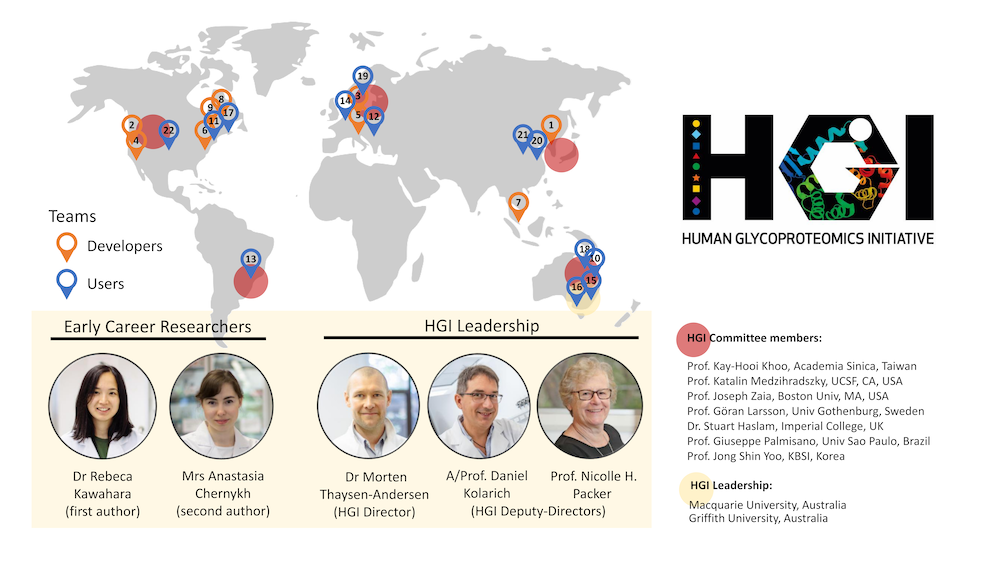
Concluding at the end of 2021, the first Human Glycoproteomics Initiative community-driven study (Kawahara et al., Nature Methods, 2021) generated many insightful conclusions and, importantly, many interesting questions about data analysis throughout the glycoproteomics community. These open questions serve as the foundation for the second HGI-led community-driven study, which is now underway and in the recruitment phase for participants. From the first publication: “Thus, a limitation of this study is that newer tools are available at the time of publication that were not compared in our analysis. Follow-up studies comparing the performance of these latest glycoproteomics software upgrades and informatics solutions not included in this study are therefore warranted. Beyond testing the ability of participants to identify the peptide and glycan components of glycopeptides from glycoproteomics data, such future comparative studies should ideally also test the ability to accurately quantify (relative, absolute) and report on modification sites of identified glycopeptides and could explore other relevant parameters not addressed herein including the use of alternative proteases, tandem mass tag-labeling and stepped-HCD-MS/MS data among other experimental conditions gaining popularity in glycoproteomics.” This second study will focus on teams of software developers only, with the goal to identify strengths and weaknesses of the very latest glycoproteomics software for glycopeptide identification and quantitation. Once all participant teams are set, sample types, data types, and reporting metrics will be decided with participant input. The deadline to join the study is May 1, 2022 If you are a glycoproteomics software tool developer and would like to be involved, please contact the co-chairs of the study. Other Community Initiatives in the Glycosciences:The Human Glycome Project | Announcements & upcoming eventsCompletion of the 1st HGI study HUPOST, 31 August 2021 27th Annual Lorne Proteomics Symposium 2022 Saturday, Feb 5 - 4:00PM Community Evaluation of Glycoproteomics Software - Morten Thaysen-Andersen Further ReadingRecent glycoproteomics reviews, non-exhaustive list... |










.png)