News in Science | HUPOST Vol. 6, Q1 Feb 2016
Kay-Hooi Khoo, Academia Sinica, Taiwan
Among the post-translational modifications, mass spectrometry (MS) analysis of protein glycosylation remains the most challenging due to the diversity in glycosylation. However, the glyco-savvy proteomic community continues to report major technical advances in all aspects of glycomics and glycoproteomics, from sample preparation, data acquisition to informatics associated with data analysis. It is particularly welcoming to see an increasing awareness and readiness to tackle analysis of intact N-glycopeptides instead of only defining the occupied N-glycosylation sites by the easier route of analyzing the de-N-glycosylated peptides. In general, the few key enabling steps in any MS-based glycoproteomic attempt to map the site-specific glycosylation pattern include 1) enrichment of the glycopeptides; 2) construction of glycan and glycosite-containing peptide library or database to allow identification of the intact glycopeptides by any of the currently available search algorithm; 3) generation of sufficiently good quality glycopeptide MS/MS data in the first place, which usually requires the presence of peptide core fragment ions, peptide core+HexNAc or the Y1 ion, and high resolution/accurate mass MS1 data of the glycopeptide precursors.
The main innovation introduced by Hui Zhang’s group at Johns Hopkins University in a recent work published in the Jan 2016 issue of Nature Biotechnology is the way the N-glycan and N-glycosite containing peptide database was generated. Most other groups resorted to identifying as many of the PNGaseF-de-N-glycosylated peptides from analysis of a separate aliquot of the enriched glycopeptide fractions and use a publicly available glycan library such as the GlycomeDB, or the default library option available in commercial software such as Byonic. The authors took a different approach by modifying their long established hydrazide chemistry-based glycopeptide-specific capture method. Solid phase immobilization was accomplished non-selectively instead by conjugation to aldehyde-functionalized solid support via reaction with the N-termini of all tryptic peptides and, after de-N-glycosylation by PNGase F, Asp-N was used to selectively cleave and release only glycosite-containing peptides with Asp at at the initially N-glycosylated Asn site. Thus only peptides carrying N-glycosites will be selectively recovered and identified. This is a very clever approach although many chemistry modification steps are involved including modifying sequentially the e-amino group of Lys and free carboxylic groups of C-termini, Asp, Glu and sialic acids. The result is quite impressive and the authors showed that it led to many more N-glycosite-containing peptides identified. The authors also went on to profile the released glycans in order to construct their own sample-specific glycan library. This was performed by both MALDI- and LC-MS and facilitated by the sialic acids now being modified by aniline. For actual analysis of the intact glycopeptides, HILIC enriched glycopeptides were subjected to LC-HCD MS/MS on a Q-Exactive instrument and the resulting spectra searched against the custom derived database using the precursor mass-matching option of their in-house developed GPQuest software. With filtering based on the presence of peptide+HexNAc and/or peptide ions, as well as ≥7 observed b and y ions (1% FDR), this resulted in positive assignment of 4,562 oxonium ion-containing spectra to 1,562 unique glycopeptides containing 518 glycosites and 81 glycans from OVCAR-3 cells.
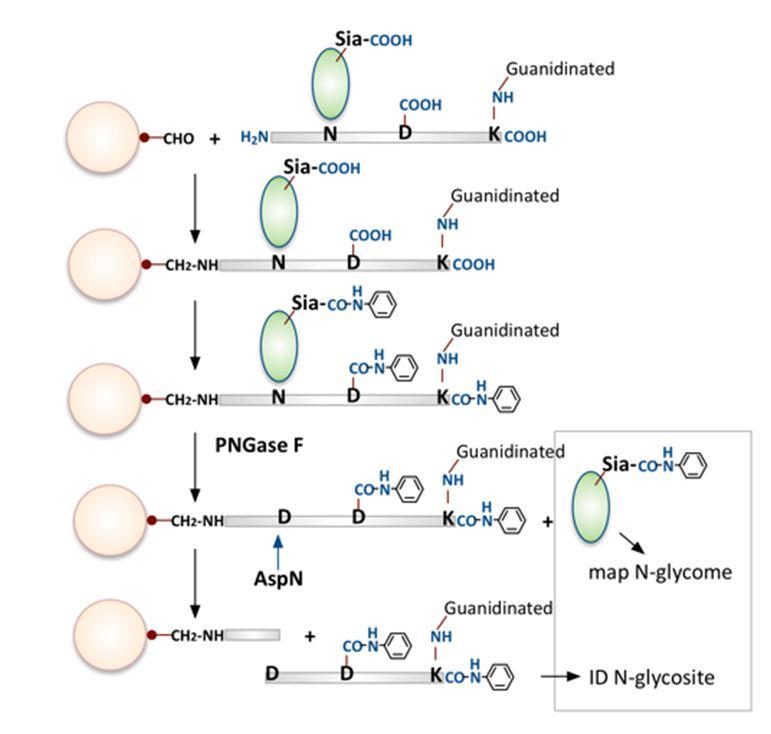
Key steps in solid phase extraction of N-glycans and N-glycosite containing peptides.
Image adapted by Dr. Kay-Hooi Khoo, Academia Sinica, Taiwan
Comprehensive analysis of protein glycosylation by solid-phase extraction of N-linked glycans and glycosite-containing peptides (2016) Sun S, Shah P, Eshghi ST, Yang W, Trikannad N, Yang S, Chen L, Aiyetan P, Hoti N, Zhang Z, Chan DW and Zhang H. Nat Biotechnol. 34, 84-88.
http://www.nature.com/nbt/journal/v34/n1/abs/nbt.3403.html


.png)