Nicki Packer, Macquarie University, Australia
Protein glycosylation is a common and diverse modification crucial to the understanding of protein function in just about every biological system. Advances in mass spectrometry now allow hundreds and even thousands of unique intact glycopeptides to be identified from a single LC-MS/MS-based glycoproteomics experiment [1]. However, confident identification of intact glycopeptides is still challenging and correct spectral annotation remain heavily dependent on laborious manual expert interpretation of fragment mass spectra. Glycoproteomics is experiencing increasing attention these years not least within the HUPO community as demonstrated with a dedicated glycoproteomics sessions at HUPO2016. However, the described technical limitations are inhibiting new scientists from entering this exciting area of proteomics. Thus, there is a clear need for tools for accurate and confident automated glycopeptide identification.
The past decade has seen promising developments of a range of MS-based software for intact glycopeptide identification [2]. To enable community evaluation of these developing tools, the new chair of the HGI, Prof Nicki Packer, Macquarie University, Sydney launched a new HGI study at HUPO2017. For this purpose, two large data files containing high resolution tandem mass spectral data of intact glycopeptides from human serum have been generated for this purpose by ThermoFisher. Combinations of dissociation modes (HCD, EThcD, ETciD and CID) were used to obtain the spectra. An expert panel will curate/analyse the data to create a consensus glycopeptide library, for comparison against the list of identifications being reported by participants in the study.
We are calling for participants (of academic and/or industrial origin) that are either developers of glycoproteomics software and/or expert users (researchers) in glycoproteomics, to identify and report on the characterization of the intact serum glycopeptides using either their own software (developers) or by using one or more tools which they routinely use in their glycoproteomics research including manual interpretation (expert users). Participants are asked to report how they interpreted the data. We plan to send out the intact glycopeptide MS/MS data this year in order to present the preliminary results of the comparative study at the next HUPO in 2018, with a planned publication of the final results and conclusions to follow. Please contact nicki.packer@mq.edu.au if you would like to be part of this study.
HGI Committee members:
Prof Nicki Packer (Macquarie University and Griffith University, Australia) (Chair)
Dr Morten Thaysen-Andersen, Macquarie University, Australia (Deputy Chair)
A/Prof Daniel Kolarich, Griffith University, Australia (Deputy Chair)
Dr Stuart Haslam, Imperial College, UK
Prof Kai-Hooi Khoo, Academia Sinica, Taiwan
Prof Goran Larson, Gothenburg University, Sweden
Prof Katalin Medzihradszky, UCSF, USA
Prof Giuseppe Palmisano, University of Sao Paulo, Brazil
Prof Jong Shin Yoo, Korea Basic Science Institute, Korea
Prof Joe Zaia, Boston University, USA
References:
[1] Thaysen-Andersen M, Packer NH, Schulz BL. Maturing Glycoproteomics Technologies Provide Unique Structural Insights into the N-glycoproteome and Its Regulation in Health and Disease. Mol Cell Proteomics. 2016. 15(6):1773.
[2] Hu H, Khatri K, Klein J, Leymarie N, Zaia J. A review of methods for interpretation of glycopeptide tandem mass spectral data. Glycoconj J. 2016. 33(3):285.
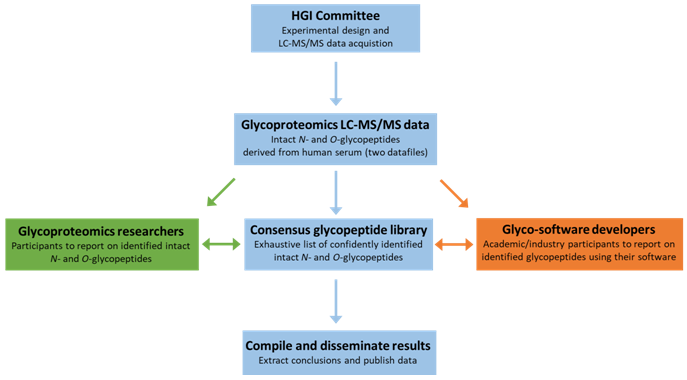
Design of the new HGI study to compare the performance of current glycoproteomics software for N- and O-glycopeptide identification from high resolution MS/MS spectral data of a complex mixture of serum proteins.


.png)