Péter Horvatovich, University of Groningen, The Netherlands
The annual meeting of the C-HPP and C-HPP PIs (PIC) was held at HUPO 2018 in Orlando, Florida. The meeting was very well attended by members and PIs. A progress report on the next-MP50 was prepared before the meeting and will be updated when all reports have been submitted and uploaded to the C-HPP Wiki. Several important decisions were made after extensive discussions. Following Young-Ki Paik’s Paik (Yonsei University) proposal to rotate the leadership of C-HPP. Chris Overall (University of British Columbia) was unanimously elected as C-HPP Chair. Young-Ki was then elected as the new Co-Chair with Lydie Lane continuing her current position as shared Co-Chair of C-HPP Executive Committee (EC).
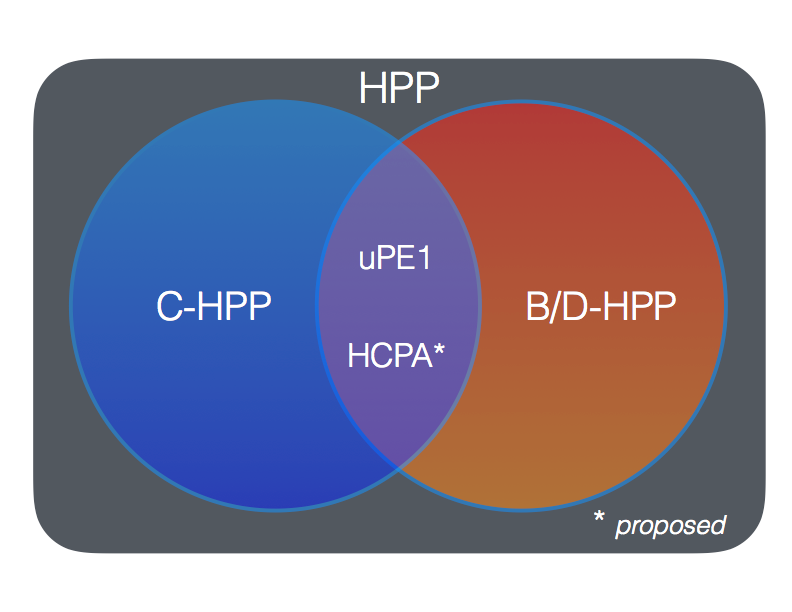 Collaborations with B/D-HPP. To further the collaborations with the B/D-HPP, the membership of the C-HPP agreed that the uPE1 project was ideal to combine with the talents and resources of the B/D-HPP, and in discussions with the B/D-HPP leadership this too was agreed (see Figure, noting that for clarity the Pillars etc. are not shown). Relevant papers in the JPR SI can be found in PMID 30269496.
Collaborations with B/D-HPP. To further the collaborations with the B/D-HPP, the membership of the C-HPP agreed that the uPE1 project was ideal to combine with the talents and resources of the B/D-HPP, and in discussions with the B/D-HPP leadership this too was agreed (see Figure, noting that for clarity the Pillars etc. are not shown). Relevant papers in the JPR SI can be found in PMID 30269496.
Human Cell Proteome Atlas. As part of the rare cells and tissues strategy to find MPs, a parallel value-added project to annotate the proteins of specific normal human cell types received considerable support with the view of creating a Human Cell Proteome Atlas that would be done in collaboration with B/D-HPP. The aim of the HCPA is to annotate the proteomes of normal human cells (fresh prepared or low passage number primary culture) so as to provide an authorative and convenient single lookup data base that would complement the other resources based on antibodies (Human Protein Atlas), Cellosaurus, and various genetic based resources. During HUPO the C-HPP initiated discussions with Dr. Neil Keller to bring this proposal to the HUPO EC for discussion and approval.
C-HPP 2.0. Deep discussions took place on a proposed “chromosome-rearrangement” of the working nucleus of the C-HPP, which had been floated to the membership at the Santiago C-HPP workshop on 23rd June. The proposed new structure, as part of “C-HPP 2.0”, would collapse the number of national Chromosome teams by approximately half based on their activities. Freed-up C-HPP team members would remain as members at large or nucleate bottom up to form new annotation C-HPP annotation teams based on protein families, cell types, or new technologies. Such teams would have equal standing with those teams wishing to remain chromosome based.
Following further discussion, the membership clearly expressed their wish and desire to pursue their national projects under the current chromosome-organised structure. Points brought up included that Chromosome teams very much liked having a recognised national “brand” as a platform to nucleate like-minded labs and centers to work collaboratively together, importantly a Chromosome “face” was valuable to interface more effectively with Universities, Funding Agencies and Governments. It was also expressed that this structure was the most effective to support the current projects, many of which are in fact chromosome orientated, i.e. finding missing proteins (neXt-MP50), discovering uPE1 protein functions (neXt-CP50), and to maintain funding and salaries by some teams that have secured significant operating funds. Members of the Italian Mt team proposed an innovative cross matrix solution that has been drawn in the figure, that was greeted with great enthusiasm as meeting the new needs and scientific objectives of the C-HPP under the HPP umbrella.
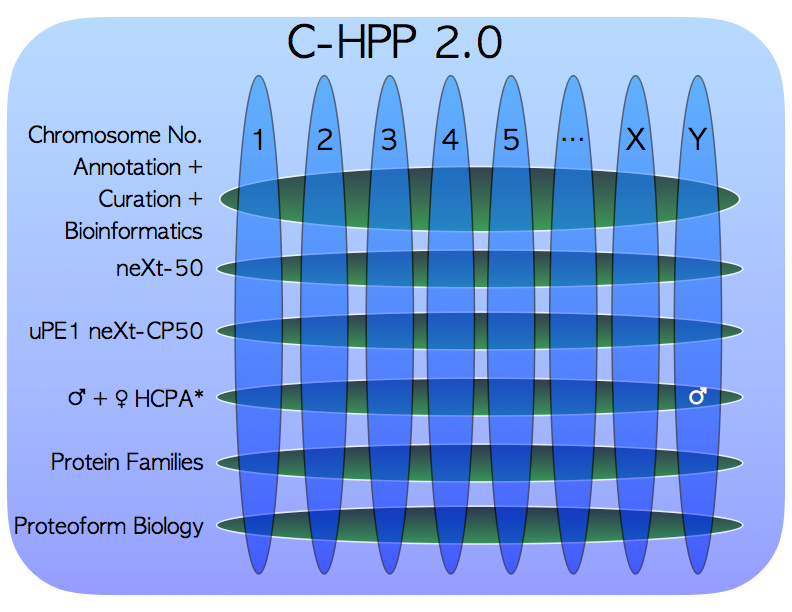
A motion to retain the existing Chromosome Structure was seconded and put to the vote, which was carried unanimously.
Bioinformatics Hub. In other activities at the bioinformatics hub shared great interactions between the leaders of the main HPP resources and tools (ProteomExchange, PetideAtlas, MASSIVE, neXtProt and HPA) in order to make the most of the available data and complete the annotation of the human proteome parts list.
Annual C-HPP Workshop. The annual C-HPP workshop entitled: “Illuminating the Dark Proteome” has a major goal to advance uPE1 functionalization in the next-CP50 project. The workshop is organized by Charles Pineau in collaboration with Chris Overall and Fernando Corrales. The workshop will take place in Saint-Malo, France, a historic city on the Brittany coast (France), from 12 to 14 May 2019, with the hotels within the actual historic citadel and featuring excellent French cuisine and wines.

HPP Scientific Terms, Definitions & Abbreviations
Definition of most often used terms such as Human Proteome Project (HPP), Chromosome-Centric Human Proteome Project (C-HPP), Biology/Disease Human Proteome Project (B/D-HPP), Protein Evidence (PE), Uncharacterized PE1 proteins (uPE1s), Missing Proteins (MPs), HPP Guidelines, Dark Proteome, Proteoforms, neXt-MP50, neXt-CP50 and Popular proteins. Exact definitions of these terms with explanation and link to scientific article as supporting reference, to additional resources such as list of MPs or uPE1s is provided in PDF document at the HUPO webpage, at the C-HPP Wiki, and as wiki page with live links at C-HPP Wiki. We encourage the scientific community to discuss further these terms and complete it with additional ones during the HUPO 2018 Orlando congress.
C-HPP Wiki update
The program of C-HPP activities at HUPO 2018 (Orlando, US, September 30-october 4, 2018) are summarized at C-HPP Wiki, showing pre and post congress C-HPP programs, Bioinformatics Hub and C-HPP sessions. Many presentations provided by authors are already available at C-HPP Wiki and others will be made public upon availability in the next few weeks. We encourage the C-HPP Chromosome members to share news, presentations and highlights on their work themselves on the C-HPP Wiki page.


.png)